RNTuple: The New Columnar Storage for HEP Data
A new data format developed by the ROOT project to address the computing challenges of HL-LHC and beyond.
25th March 2025 | Source: CERN EP newsletter
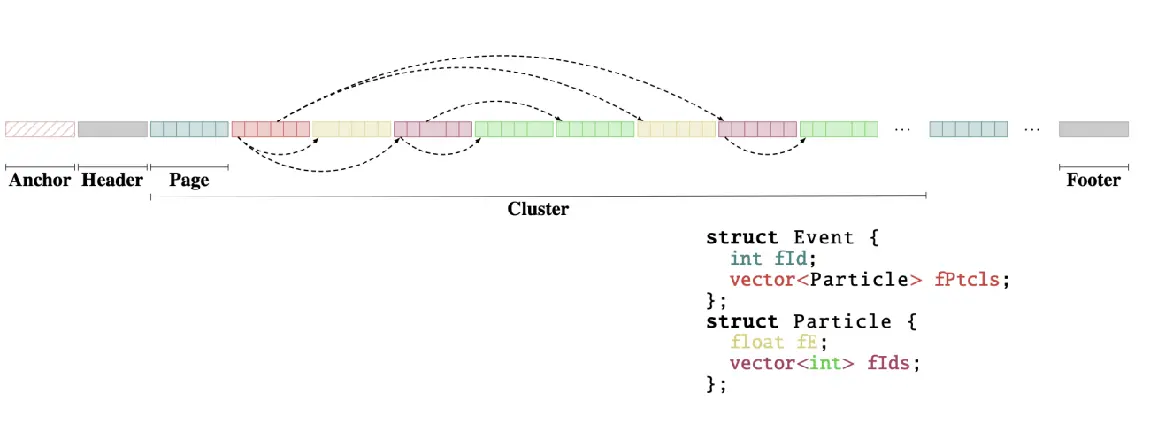
R&D activities are vital for the ROOT project. A few years ago, we started investigating the evolution of TTree, ROOT’s columnar format with which more than two exabytes of data were written at the LHC: experiments write data, lots of it, to then store them on media such as disk or tape. A storage challenge is ahead of us in the HL-LHC era, during which experiments will acquire about ten times more data than the data collected by the end of Run 3.
After 6 years of R&D, RNTuple reached an important milestone: we released the first official version of the on-disk binary format! The first ROOT version where this format is honoured is ROOT 6.34, released in November 2024. What does it mean for you, in a nutshell? It means that all future ROOT versions will be able to read back such objects. That may sound trivial, yet it has significant implications behind the scenes. The on-disk format, to recap, defines the layout of the bits on storage media that hold a certain data set. Once the format is fixed and applications write (large amounts of) data, there are severe restrictions on what can still be practically changed.
Why then, you may ask, did we change at all from the TTree format? It was working very well after all, right? The reason is that some of RNTuple’s biggest advantages are intimately linked to the on-disk representation. Perhaps most importantly: the savings in data volume. For instance, for both CMS NanoAODs and ATLAS PHYSLITE files, compared to TTree we can store the same content in substantially fewer bytes:
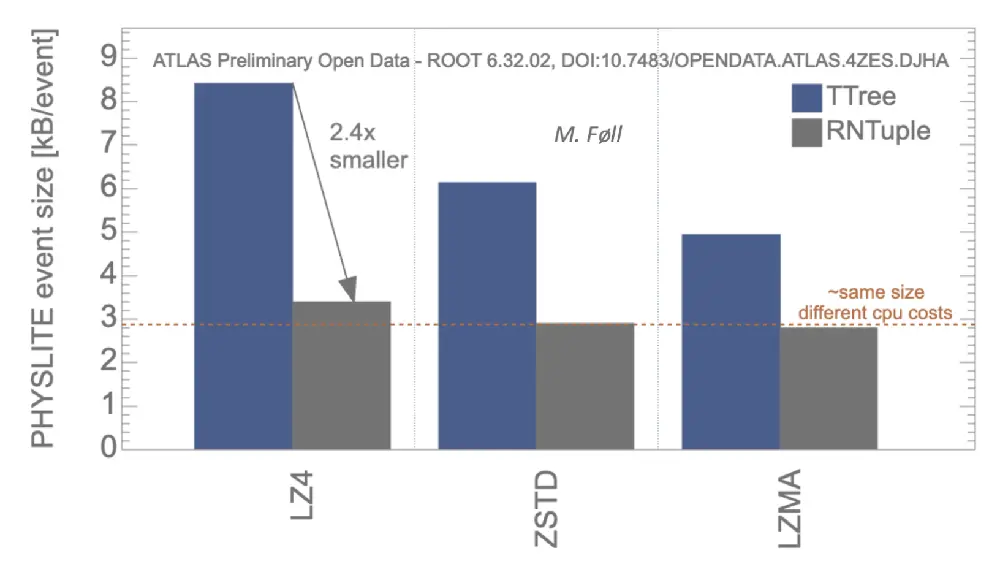
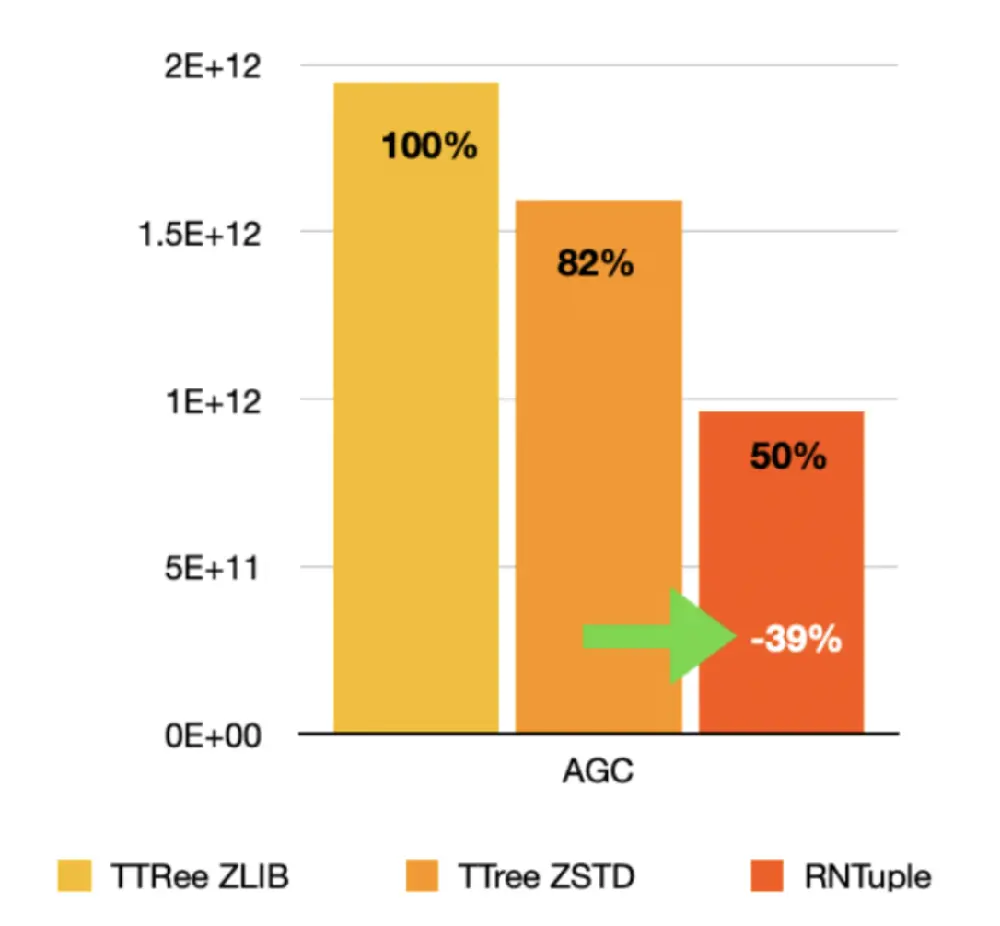
This, and other reasons related to the robustness and the use of modern storage systems, convinced us to introduce a new on-disk format after more than 25 years of TTree. With RNTuple, we aim at a new, stable format for the exabytes of data to be expected during the lifetime of upcoming experiments (e.g., the experiments at the HL-LHC, EIC, DUNE, and beyond).



